Deep Learning Approaches for Predicting Glaucoma Progression Requiring Surgery Using Electronic Health Records Data and Natural Language Processing
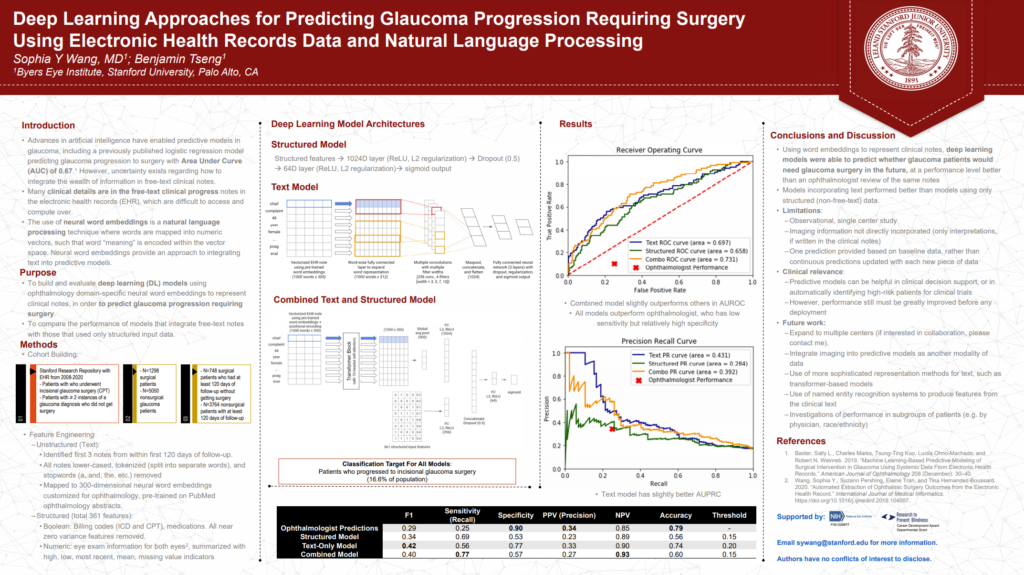
Some of my work with the Stanford Ophthalmic Informatics and Artificial Intelligence Group recently made it into a poster presentation at 2021 (virtual) meeting of the American Glaucoma Society. It also made it into a published paper in Ophthalmology Science: “Deep Learning Approaches for Predicting Glaucoma Progression Using Electronic Health Records and Natural Language Processing” [PDF]
In it, we described some of our work applying deep learning methods to predicting whether or not a given glaucoma patient will advance to requiring surgery. This is a noteworthy problem as, given that glaucoma is a chronic disease with no present cure, being able to understand which patients are at greatest risk of escalation may better help clinicians prioritize intervention and follow-up. This is also an especially challenging diagnostic problem as there are numerous factors, both known and unknown, which contribute to glaucoma progression.
While there have been machine learning models built capturing detailed structured data (i.e. objective fields like intraocular pressure, etc.), there has been little done to study the predictive value of the unstructured data (i.e. clinician notes in electronic medical record).
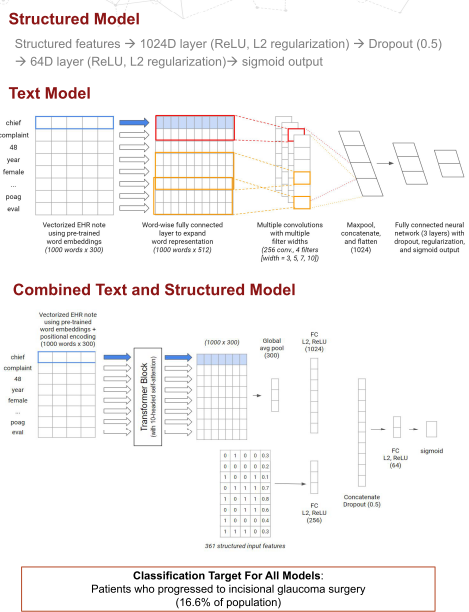
We developed an unstructured model and a combined structure + unstructured model which took data from a large corpus of ophthalmology EHR notes from Stanford’s STARR (Stanford Research Repository) dataset and applied transfer learning using word embeddings trained on published abstracts in medical literature on ophthalmology to make predictions on glaucoma progression following a medical visit. Structured data included billing codes, medications, and eye exam data, while unstructured text included the complete medical record within 120 days of follow-up.
We took advantage of a textCNN architecture that served us well previously in a low vision accuity prognostic task for the pure unstructured model and a combination model which took advantage of the Transformer architecture.
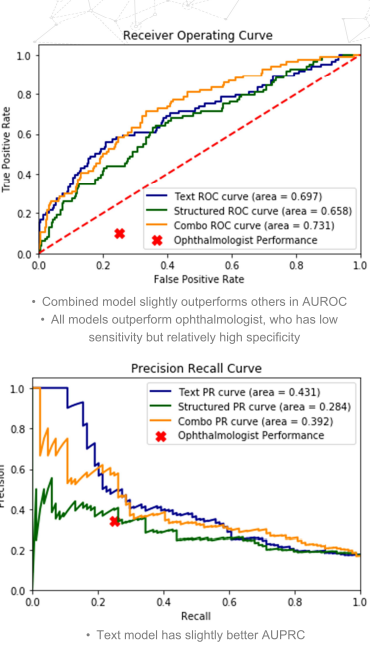
Both the unstructured (text)-only model and the combination model did meaningfully better than the best pure structured model we were able to train when measured using AUROC (0.697 and 0.731, respectively vs. 0.658), AUPRC (0.431 and 0.392, respectively vs 0.284), and F1 (0.42 and 0.40, respectively vs 0.34), as well as in comparison with a trained ophthalmologist (F1: 0.29).
Future work will include expanding the dataset beyond a single center and moving this to making continuous predictions rather than a basic one just using baseline data. There will also be additional work on model architecture using more sophisticated Transformer-based models as well as new feature-extraction taking advantage of the inherent structure to EMR notes as well as named entity recognition.
