Evaluating Ophthalmology-Specific Word Embeddings Using Analogies and Low Vision Prognosis Prediction
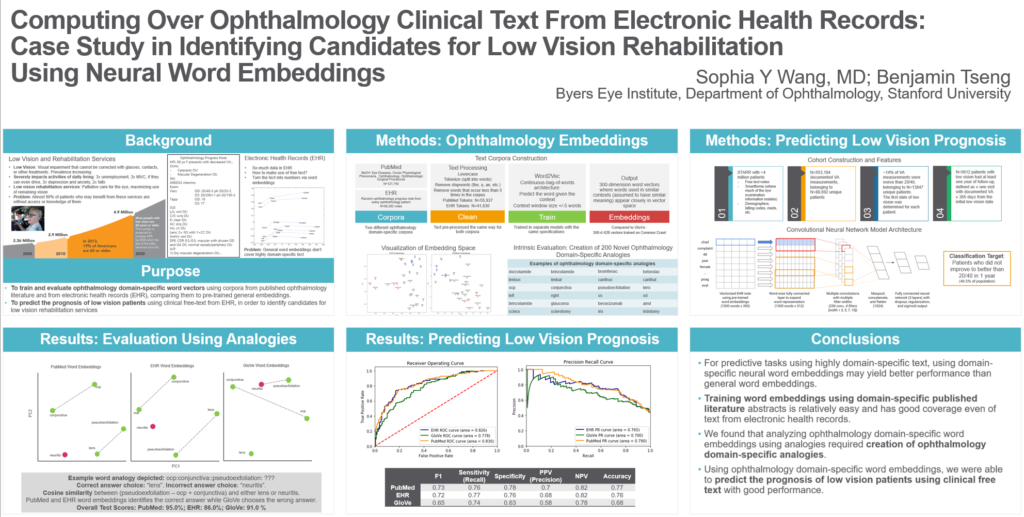
Some of my work with Professor Sophia Wang of Stanford funded by Research to Prevent Blindness was presented in a poster presentation at the 2020 Mobilizing Computable Biomedical Knowledge meeting. It also made it into two published papers in the International Journal of Medical Informatics: “Development and Evaluation of novel ophthalmology domain-specific neural word embeddings to predict visual prognosis”[PDF] and “Looking for low vision: Predicting visual prognosis by fusing structured and free-text data from electronic health records”[PDF]
In it, we described some of our work applying deep learning methods to the problem of interpreting doctors’ notes in electronic health records (EHRs). Specifically, we profiled the quality of word embeddings — mathematical representations of words that can be interpreted by a computer and, more importantly, can be “pre-trained” by feeding it different sources of input which allows meanings to be first learned from a body of text before being applied to another problem. In this study, we looked at three different sources of input text: abstracts from published articles in the medical literature pertaining to ophthalmology (the medical specialty of focus for this study), a large corpus of ophthalmology EHR notes from STARR (Stanford Research Repository), and the general internet (here, we specifically used pre-trained GLoVe vectors).
These word embeddings were compared on two tasks:
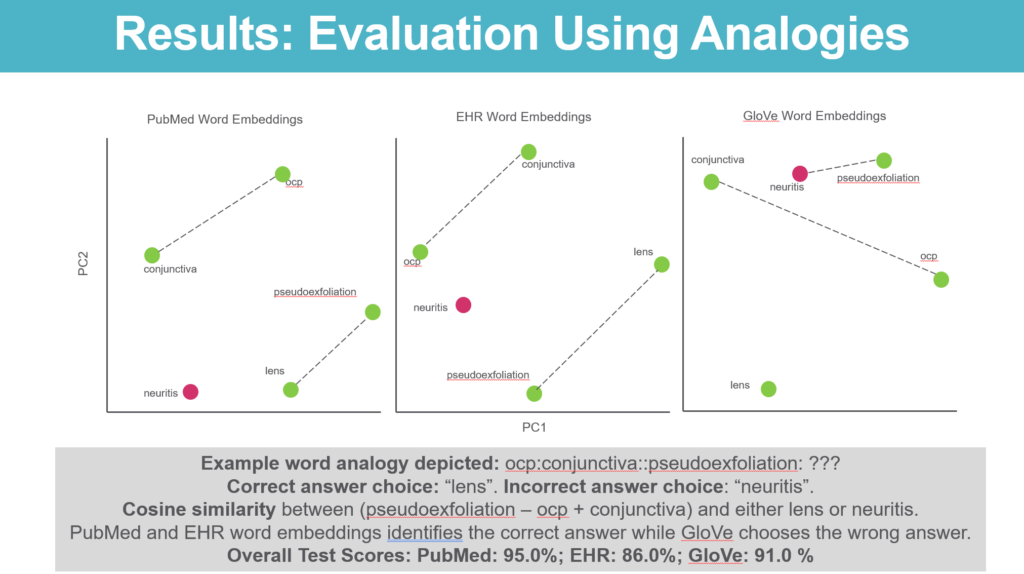
- Ophthalmology analogies – We developed a novel set of 200 ophthalmology-field specific analogies (for example: brinzolamide:glaucoma::bevacizumab:amd — an analogy pairing a drug with the condition it can treat) and tested which set of embeddings were better able to choose between the correct answer and an incorrect one. One challenge with this exercise was making sure we designed analogies using words that were found in all 3 datasets (to be able to compare results — in practice there were many jargons and acronyms that are covered in the EHR text and ophthalmology literature abstracts that aren’t in the general internet text corpus)
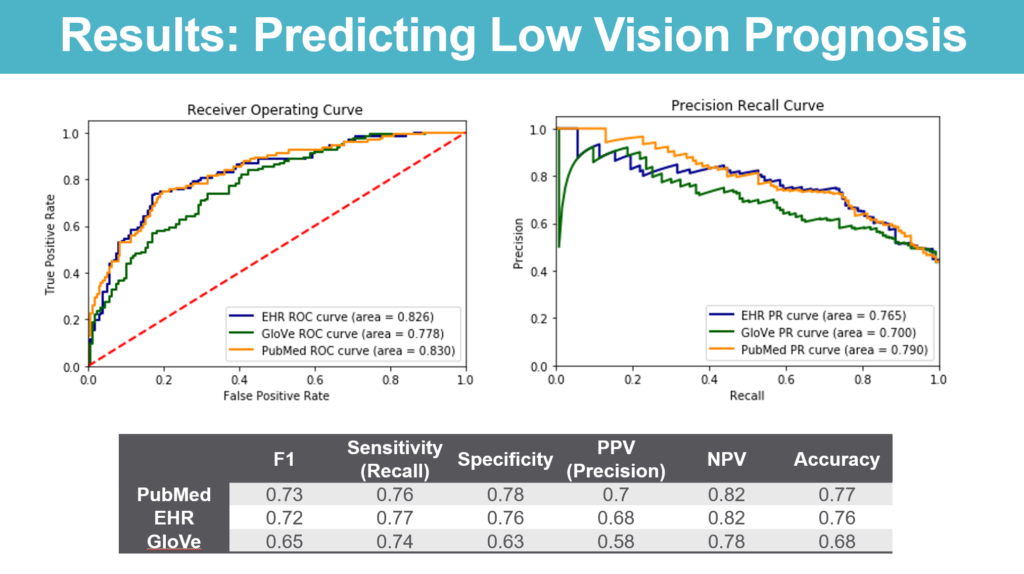
- Predicting prognosis of low vision patients – We used STARR to identify patients who were diagnosed with low vision with at least an additional followup point at least a year later and looked at the performance of deep learning models trained using the different types of embeddings in predicting whether or not these patients continued to be low vision at the followup.
We hypothesized that word embeddings developed using general internet text would do less well on these tasks as general internet text would be unlikely to encode the meaning of ophthalmology-specific jargon and, hence would be less able to interpret the medical context needed on the analogies or make accurate predictions on disease prognosis. I personally also hypothesized that, while the embeddings trained on ophthalmology literature would outperform embeddings from the general internet, that they would underperform embeddings that were trained on EHR notes as the way physicians write clinical notes is very different from how they write journal articles.
Interestingly enough, we found that the embeddings trained on ophthalmology published literature abstracts did the best on analogies (95% correct) and, interestingly enough, the embeddings trained on EHR text actually did the worst (86% correct) with the embeddings trained using the general internet (GLoVe vectors) doing somewhere in between (91% correct).
On the other hand, models to predict disease progression taking embeddings trained using the ophthalmology literature or the EHR performed similarly while outperforming models trained using general-purpose word embeddings.
There will be more work to come here — but the analysis shows how AI/NLP performance on specialized tasks can be improved by taking advantage of subject-specific bodies of text. It also bodes well for future work developing more sophisticated models to tackle medically important problems such as identifying patients to prioritize for interventions like low vision rehabilitation.
The published paper can be found in the International Journal of Medical Informatics; https://doi.org/10.1016/j.ijmedinf.2021.104464
