This month, I read a paper (HT: my ex-college roommate Eric) by a group from Beth Israel about systemic inflammatory response syndrome (SIRS) following serious injury. SIRS, which is more commonly understood/found as sepsis, happens when the entire body is on high “immune alert.” In the case of sepsis, this is usually due to an infection of some sort. While an immune response may be needed to control an internal infection, SIRS is dangerous because the immune system can cause a great deal of collateral damage, resulting in potentially organ failure and death.
Whereas an infection has a clear link to sepsis, the logic for why injury would cause a similar immune response was less clear. In fact, for years, the best hypothesis from the medical community was that injury would somehow cause the bacteria which naturally live in your gut to appear where they’re not supposed to be. But this explanation was not especially convincing, especially in light of injuries like burns which could still lead to SIRS but which didn’t seem to directly affect gut bacteria.
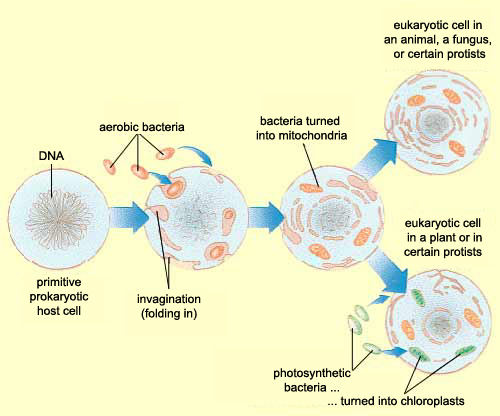
Zhang et al, instead of assuming that some type of endogenous bacteria was being released following injury, came up with an interesting hypothesis: it’s not bacteria which is triggering SIRS, but mitochondria. A first year cell biology student will be able to tell you that mitochondria are the parts of eukaryotic cells (sophisticated cells with nuclei) which are responsible for keeping the cell supplied with energy. A long-standing theory in the life science community (pictured above) is that mitochondria, billions of years ago, were originally bacteria which other, larger bacteria swallowed whole. Over countless rounds of evolution, these smaller bacteria became symbiotic with their “neighbor” and eventually adapted to servicing the larger cell’s energy needs. Despite this evolution, mitochondria have not lost all of their (theorized) bacterial ancestry, and in fact still retain bacteria-like DNA and structures. Zhang et al’s guess was that serious injuries could expose a mitochondria’s hidden bacterial nature to the immune system, and cause the body to trigger SIRS as a response.
Interesting idea, but how do you prove it? The researchers were able to show that 15 major trauma patients with no open wounds or injuries to the gut had thousands of times more mitochondrial DNA in their bloodstream than non-trauma victims. The researchers were then able to show that this mitochondrial DNA was capable of activating polymorphonuclear neutrophils, some of the body’s key “soldier” cells responsible for causing SIRS.
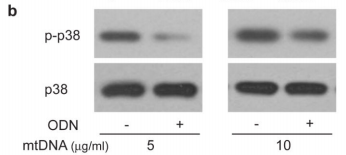
The figure above shows the result of an experiments illustrating this effect looking at the levels of a protein called p38 MAPK which gets chemically modified into “p-p38” when neutrophils are activated. As you can see in the p-p38 row, adding more mitochondrial DNA (mtDNA, “-” columns) to a sample of neutrophils increases levels of p-p38 (bigger, darker splotch), but adding special DNA which blocks the neutrophil’s mtDNA “detectors” (ODN, “+” columns) seems to lower it again. Comparing this with the control p38 row right underneath shows that the increase in p-p38 is likely due to neutrophil activation from the cells detecting mitochondrial DNA, and not just because the sample had more neutrophils/more p38 (as the splotches in the second row are all roughly the same).
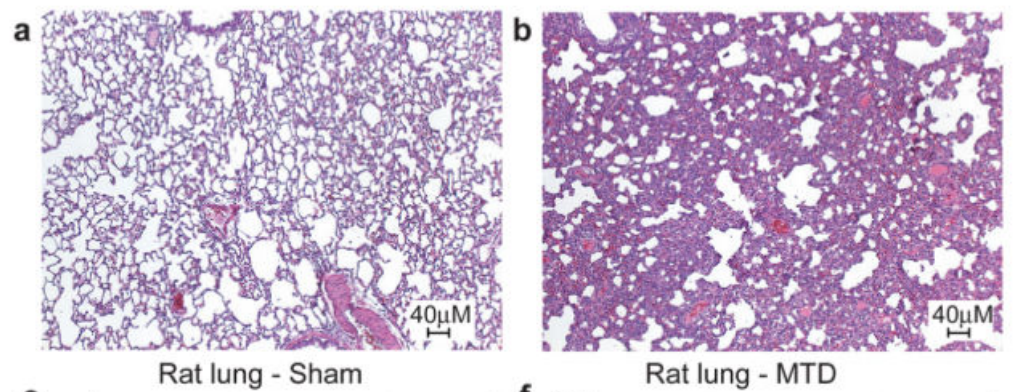
Cool, but does this mean that mitochondrial DNA actually causes a strong immune response outside of a test tube environment? To test this, the researchers injected mitochondrial DNA into rats and ran a full set of screens on them. While the paper showed numerous charts pointing out how the injected rats had strong immune response across multiple organs, the most striking are the pictures below which show a cross-section of a rat’s lungs comparing rats injected with a buffer solution (panel a, “Sham”) and rats injected with mitochondrial DNA (panel b, MTD). The cross-sections are stained with hematoxylin and eosin which highlight the presence of cells. The darker and “thicker” color on the right shows that there are many more cells in the lungs of rats injected with mitochondrial DNA – most likely from neutrophils and other “soldier cells” which have rushed in looking for bacteria to fight.
Amazing isn’t it? Not only did they provide part of the solution to the puzzle of injury-mediated SIRS (what they used to call “sterile SIRS”), but lent some support to the endosymbiont hypothesis!
Paper: Zhang, Qin et al. “Circulating Mitochondrial DAMPs Cause Inflammatory Responses to Injury.” Nature 464, 104-108 (4 March 2010) – doi:10.1038/nature08780
