I’m pretty late for paper of the month, so here we go
“Omics” is the hot buzz-suffix in the life sciences for anything which uses the new sequencing/array technologies we now have available. You don’t study genes anymore, you study genomics. You don’t study proteins anymore – that’s so last century, you study proteomics now. And, who studies metabolism? Its all about metabolomics. There’s even a blog covering this with the semi-irreverent name “Omics! Omics!”.
This month’s paper from Science is from researchers at the NIH because it was the first time I ever encountered the term “antibodyome”. As some of you know, antibodies are the “smart missiles” of your immune system – they are built to recognize and attack only one specific target (i.e. a particular protein on a bacteria/virus). This ability is so remarkable that, rather than rely on human-generated constructs, researchers and biotech companies oftentimes choose to use antibodies to make research tools (i.e. using fluorescent antibodies to label specific things) and therapies (i.e. using antibodies to proteins associated with cancer as anti-cancer drugs).
How the immune system does this is a fascinating story in and of itself. In a process called V(D)J recombination – the basic idea is that your immune system’s B-cells mix, match, and scramble certain pieces of your genetic code to try to produce a wide range of antibodies to hit potentially every structure they could conceivably see. And, once they see something which “kind of sticks”, they undergo a process called affinity maturation to introduce all sorts of mutations in the hopes that you create an even better antibody.
Which brings us to the paper I picked – the researchers analyzed a couple of particularly effective antibodies targeted at HIV, the virus which causes AIDS. What they found was that these antibodies all bound the same part of the HIV virus, but when they took a closer look at the 3D structures/the B-cell genetic code which made them, they found that the antibodies were quite different from one another (see Figure 3C below)
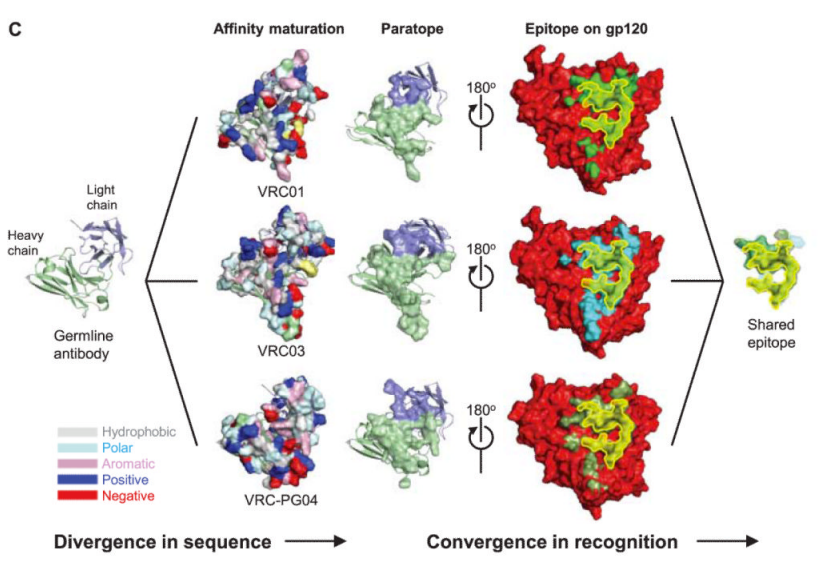
What’s more, not only were they fairly distinct from one another, they each showed *significant* affinity maturation – while a typical antibody has 5-15% of their underlying genetic code modified, these antibodies had 20-50%! To get to the bottom of this, the researchers looked at all the antibodies they could pull from the patient – their “antibodyome” (in the same way that a patient’s genome would be all of their genes) — and along with data from other patients, they were able to construct a genetic “family tree” for these antibodies (see Figure 6C below)
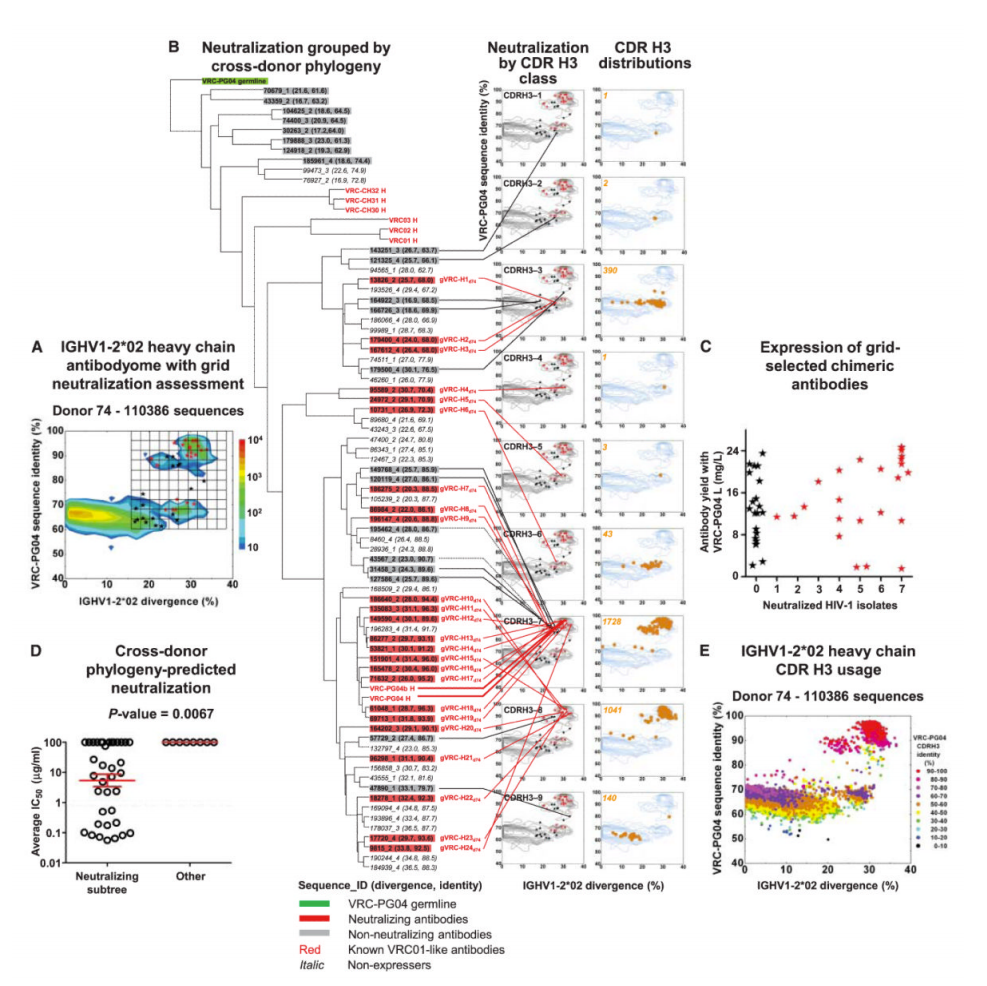
The analysis shows that many of the antibodies were derived from the same initial genetic VDJ “mix-and-match” but that afterwards, there were quite a number of changes made to that code to get the situation where a diverse set of structures/genetic codes could attack the same spot on the HIV virus.
While I wish the paper probed deeper into actual experimentation to take this analysis further (i.e. artificially using this method to create other antibodies with similar behavior), this paper goes a long way into establishing an early picture of what “antibodyomics” is. Rather than study the total impact of an immune response or just the immune capabilities of one particular B-cell/antibody, this sort of genetic approach lets researchers get a very detailed, albeit comprehensive look at where the body’s antibodies are coming from. Hopefully, longer term this also turns into a way for researchers to make better vaccines.
Paper: Wu et al., “Focused Evolution of HIV-1 Neutralizing Antibodies Revealed by Structures and Deep Sequencing.” Science (333). 16 Sep 2011. doi: 10.1126/science.1207532

Leave a Reply