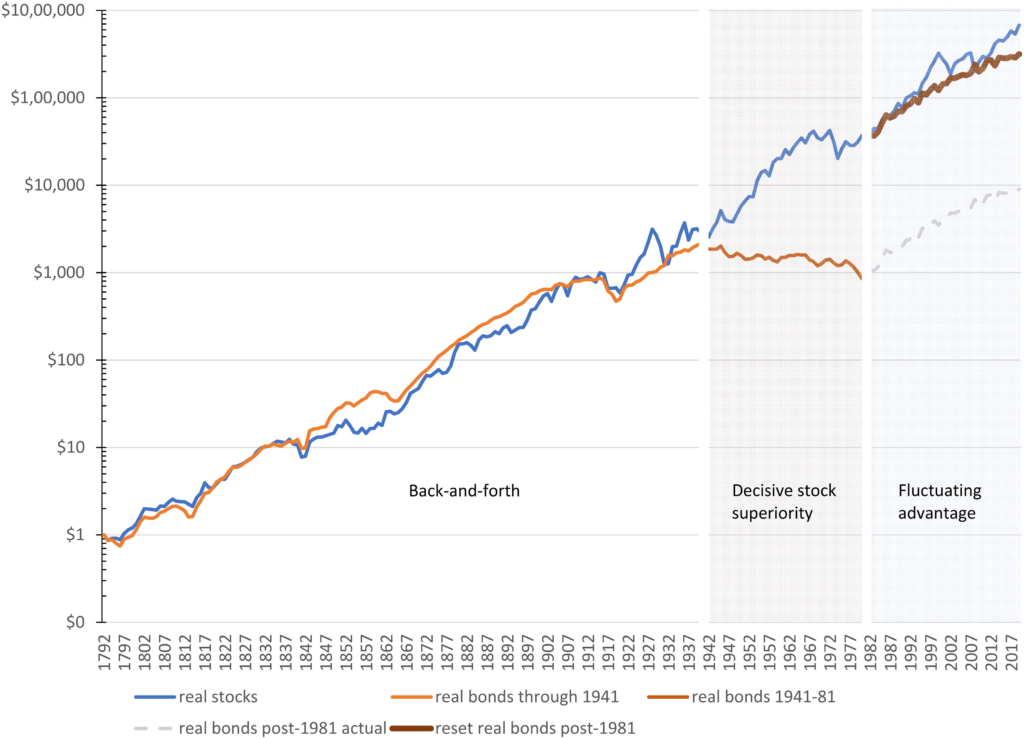
A recent preprint from Stanford has demonstrated something remarkable: AI agents working together as a team solving a complex scientific challenge.
While much of the AI discourse focuses on how individual large language models (LLMs) compare to humans, much of human work today is a team effort, and the right question is less “can this LLM do better than a single human on a task” and more “what is the best team-up of AI and human to achieve a goal?” What is fascinating about this paper is that it looks at it from the perspective of “what can a team of AI agents achieve?”
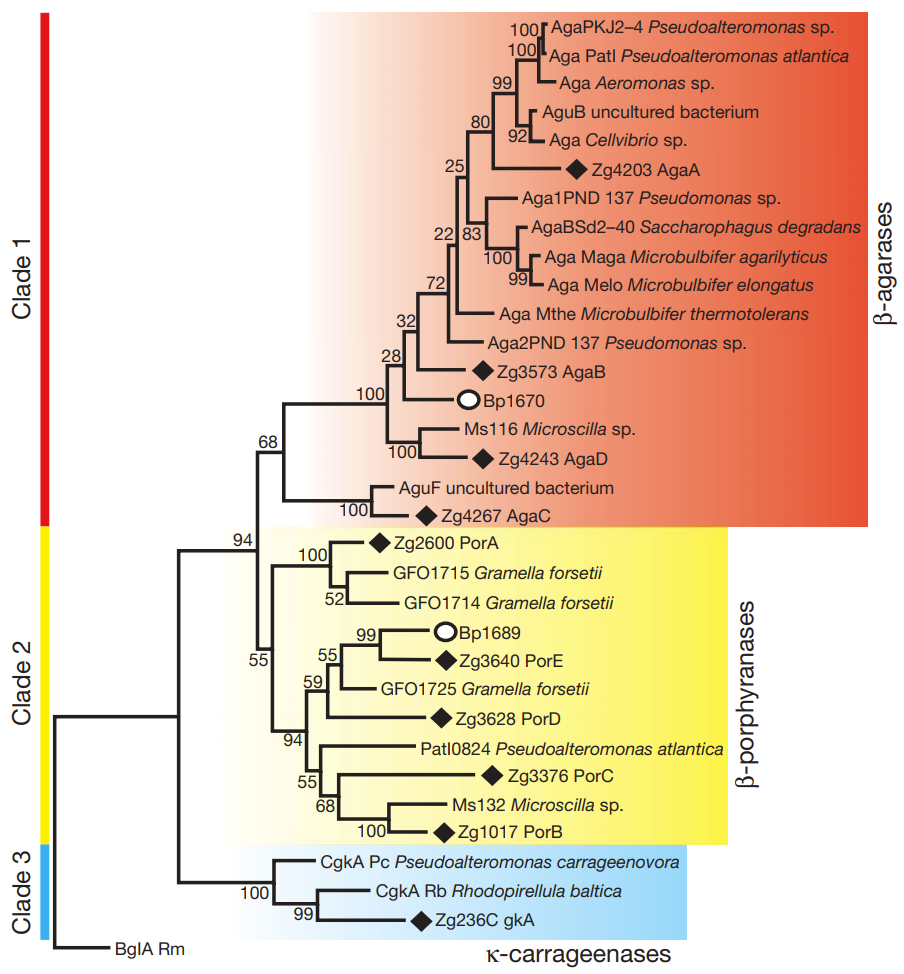
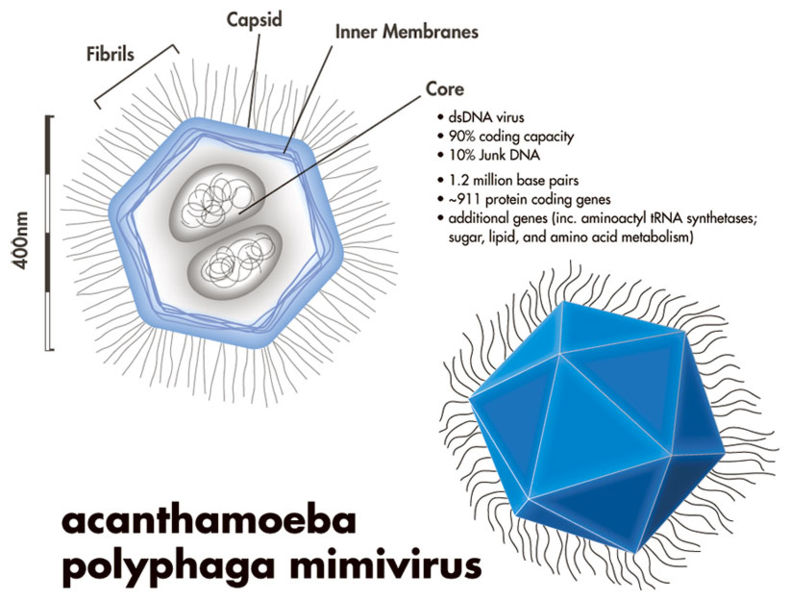
The researchers tackled an ambitious goal: designing improved COVID-binding proteins for potential diagnostic or therapeutic use. Rather than relying on a single AI model to handle everything, the researchers tasked an AI “Principal Investigator” with assembling a virtual research team of AI agents! After some internal deliberation, the AI Principal Investigator selected an AI immunologist, an AI machine learning specialist, and an AI computational biologist. The researchers made sure to add an additional role, one of a “scientific critic” to help ground and challenge the virtual lab team’s thinking.
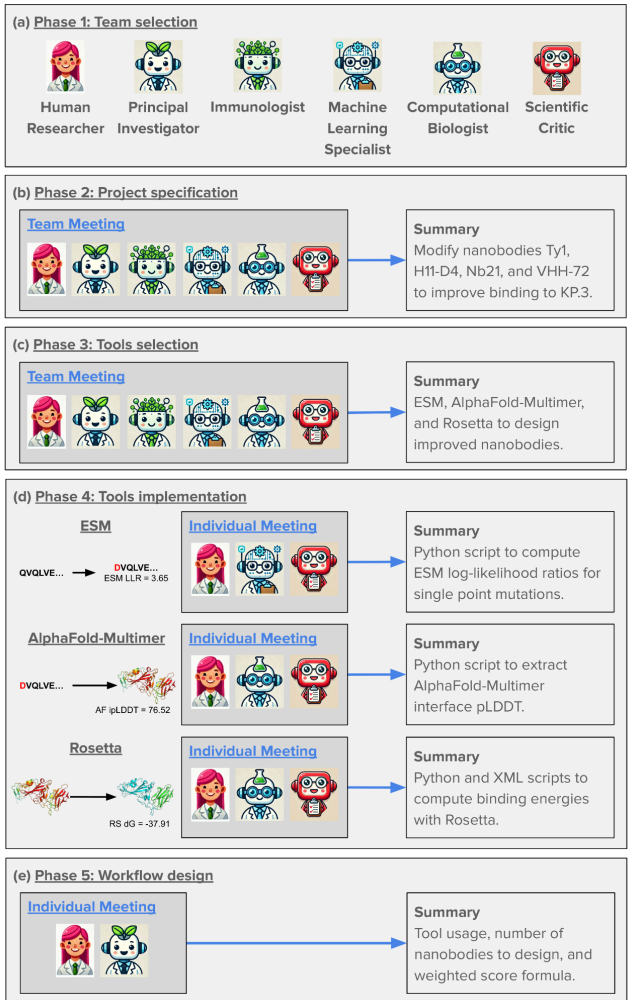
(Source: Figure 2 from Swanson et al.)
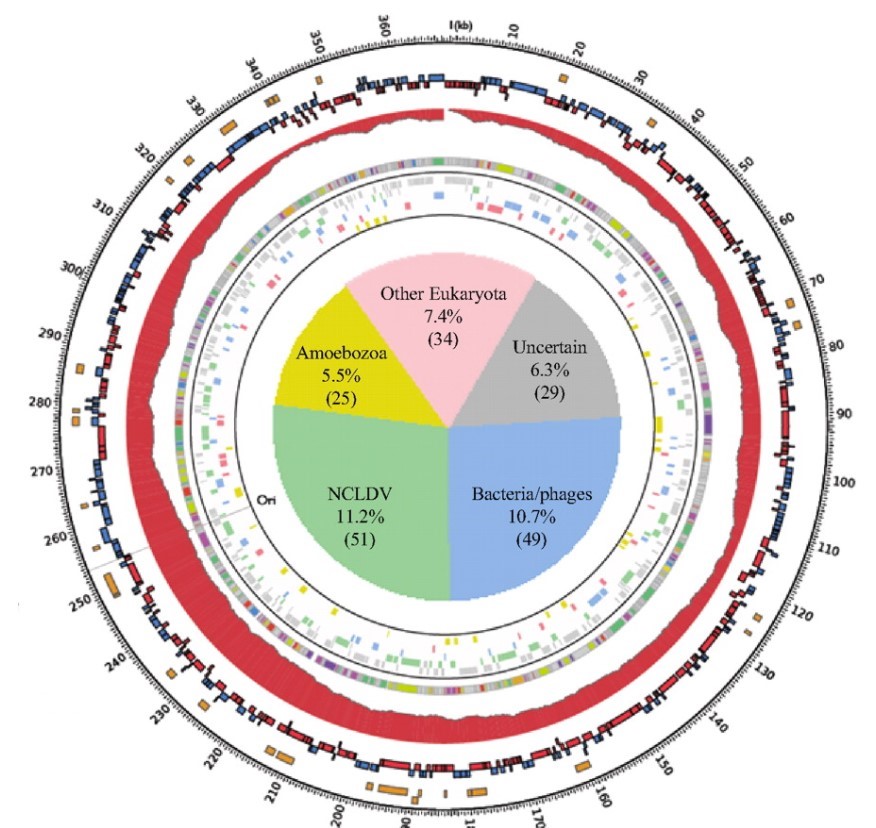
What makes this approach fascinating is how it mirrors high functioning human organizational structures. The AI team conducted meetings with defined agendas and speaking orders, with a “devil’s advocate” to ensure the ideas were grounded and rigorous.

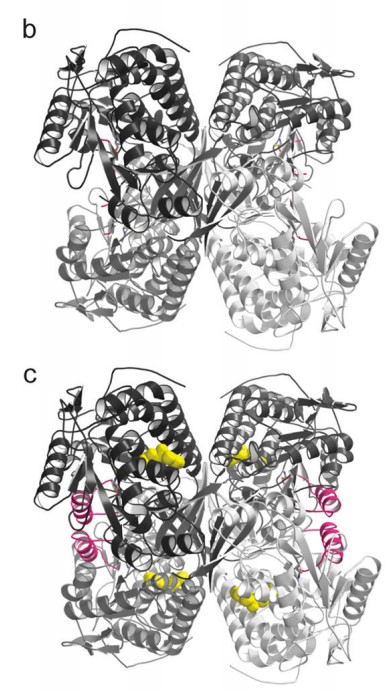
(Source: Figure 6 from Swanson et al.)
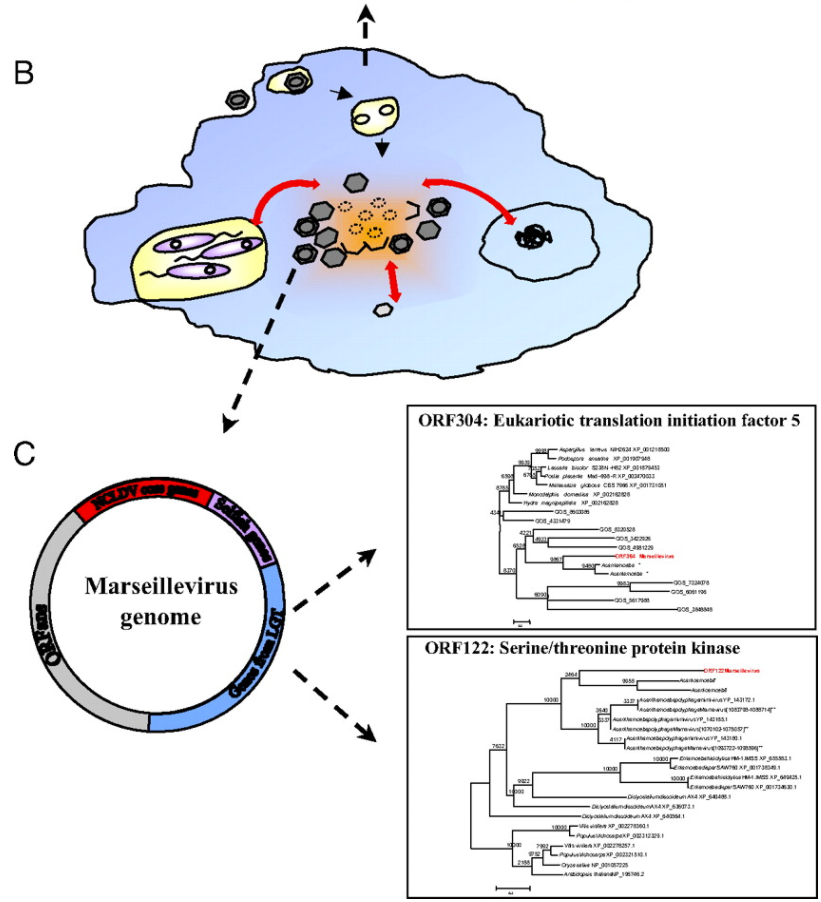
One tactic that the researchers said helped with boosting creativity that is harder to replicate with humans is running parallel discussions, whereby the AI agents had the same conversation over and over again. In these discussions, the human researchers set the “temperature” of the LLM higher (inviting more variation in output). The AI principal investigator then took the output of all of these conversations and synthesized them into a final answer (this time with the LLM temperature set lower, to reduce the variability and “imaginativeness” of the answer).

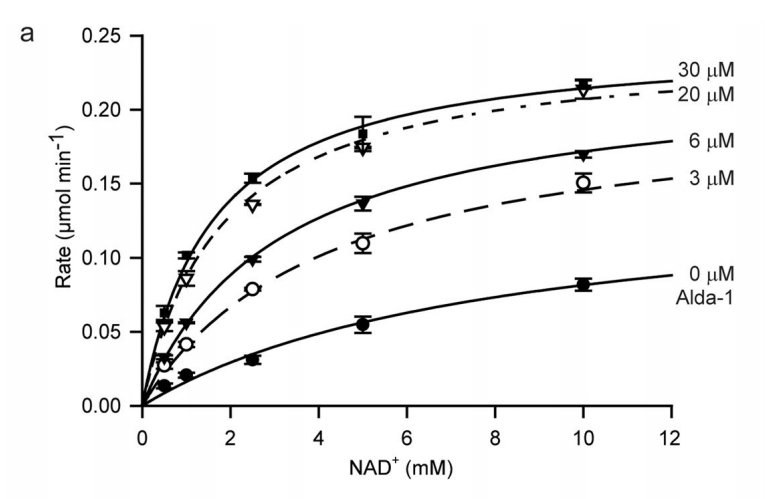
(Source: Supplemental Figure 1 from Swanson et al.)
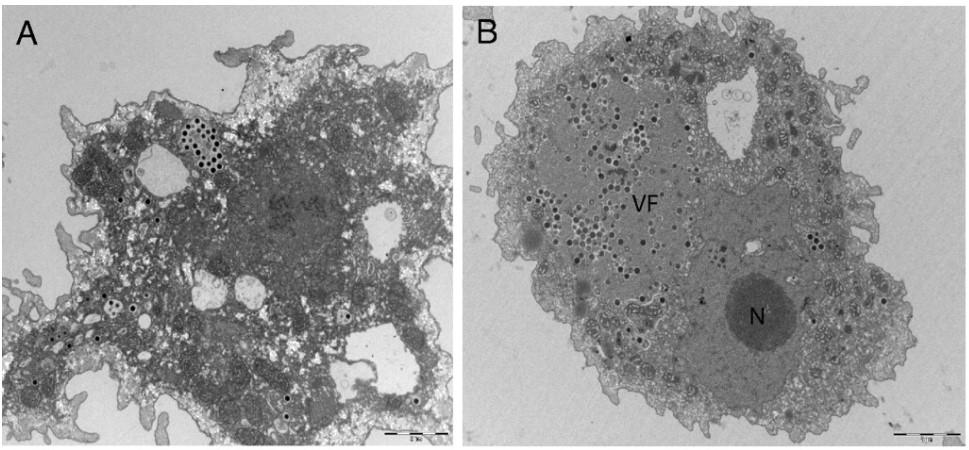
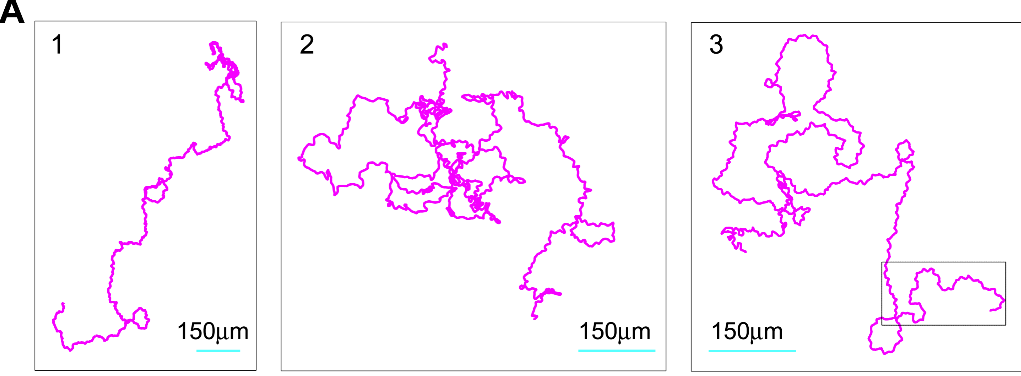
The results? The AI team successfully designed nanobodies (small antibody-like proteins — this was a choice the team made to pursue nanobodies over more traditional antibodies) that showed improved binding to recent SARS-CoV-2 variants compared to existing versions. While humans provided some guidance, particularly around defining coding tasks, the AI agents handled the bulk of the scientific discussion and iteration.

(Source: Figure 5C from Swanson et al.)
This work hints at a future where AI teams become powerful tools for human researchers and organizations. Instead of asking “Will AI replace humans?”, we should be asking “How can humans best orchestrate teams of specialized AI agents to solve complex problems?”
The implications extend far beyond scientific research. As businesses grapple with implementing AI, this study suggests that success might lie not in deploying a single, all-powerful AI system, but in thoughtfully combining specialized AI agents with human oversight. It’s a reminder that in both human and artificial intelligence, teamwork often trumps individual brilliance.
I personally am also interested in how different team compositions and working practices might lead to better or worse outcomes — for both AI teams and human teams. Should we have one scientific critic, or should their be specialist critics for each task? How important was the speaking order? What if the group came up with their own agendas? What if there were two principal investigators with different strengths?
The next frontier in AI might not be building bigger models, but building better teams.

The Virtual Lab: AI Agents Design New SARS-CoV-2 Nanobodies with Experimental Validation
Kyle Swanson et al. | bioRxiv PrePrint